
The fight against SARS-CoV-2 is one of the greatest examples of scientific collaboration ever seen. Scientists across the globe are working together to combat an invisible threat, with an effective vaccine created in less than a year.
A team led by Professor Sean O’Donoghue, from the Garvan Institute of Medical Research and CSIRO’s Data61, has produced a resource that can assist researchers with collaboration – and also with the virus’s invisibility. They’ve recently published a paper in Molecular Systems Biology[Link will open in a new window] detailing what they’ve done.
The team has compiled a library of more than 2,000 different structures involving COVID-19’s 27 proteins. The work identified viral proteins that ‘mimic’ and ‘hijack’ human proteins – tactics that allow the virus to bypass cell defences and replicate.
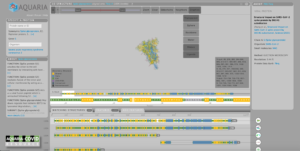
Any researcher with an internet connection can freely access these structural models on the Aquaria-COVID resource[Link will open in a new window], a website designed by the team to help the research community ‘zoom in’ on potential new targets on the virus for future treatments or vaccines, and crucially, investigate new virus variants.
The replication ‘strategy’ of viruses uses a combination of stealth, deception and cellular-level piracy.
Many viruses present a bland exterior that doesn’t trigger the target cell’s defences. Inside, however, are all the instructions, in the form of DNA or RNA, needed to take over the host cell and to build new viral copies. This process of host cell takeover is known as ‘molecular hijacking.’ When a virus replicates it must build entire viral particles and it has to not only make viral proteins, but it must also replicate its genetic material.
To better understand the biological processes used in viral replication, researchers determine the 3D shape of individual proteins.
Professor O’Donoghue and his team modelled 3D structures of all SARS-CoV-2 proteins, and generated 2,060 models that cover 69 per cent of the viral proteome[Link will open in a new window], and provide details not available elsewhere.
They found that around 6 per cent of SARS-CoV-2’s total proteins are involved in mimicking human proteins, and about 7 per cent are implicated in a range of hijacking processes. Approximately 29 per cent self-assemble into multi-unit states that are likely integral to the way the combination of viral replication and translation forms.
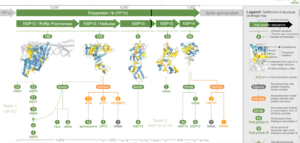
The team followed that up by making the 3D models more accessible, devising a structural coverage map, a novel visualisation method to show what is, and equally importantly, is not, known about the 3D structure of the viral proteome.
This is all contained within the Aquaria-COVID resource[Link will open in a new window]. Scientists can use emerging structural data to understand the mechanisms that underpin COVID-19 infection, or concentrate on the 31 per cent of the virus’s proteome still either structurally unknown or dark.
“Thanks to a recent research focus on SARS-CoV-2, scientists have determined around a thousand 3D structures of the virus’s 27 individual proteins, and nearly a thousand more for related proteins,” explains Professor O’Donoghue. “However, until now there has been no easy way to bring all the pieces of data together and analyse them.
“Our resource contains a level of detail of SARS-CoV-2’s structure not available anywhere else. This has given us an unprecedented insight into the virus’s activity.”
“Our analysis has highlighted key mechanisms used by the coronavirus, and these mechanisms, in turn, may guide the development of new therapies and vaccines.”
The team’s analysis reveals opportunities for further research. “Much of the coronavirus research to date has focused on the spike glycoprotein, which is the main target for current vaccines. This protein will continue to be an important target, but it’s also important we broaden our focus to other potential targets and better understand the entire viral lifecycle,” says Professor O’Donoghue.
He adds that the Aquaria-COVID resource may help researchers more easily investigate how new variants of coronavirus differ – and critically, how they may better be targeted with vaccines and treatments.
“The longer the virus circulates, the more chances it has to mutate and form new variants such as the Delta strain,” says Professor O’Donoghue. “Our resource will help researchers understand how new strains of the virus differ from each other – a piece of the puzzle that we hope will help in dealing with new variants as they emerge.”
